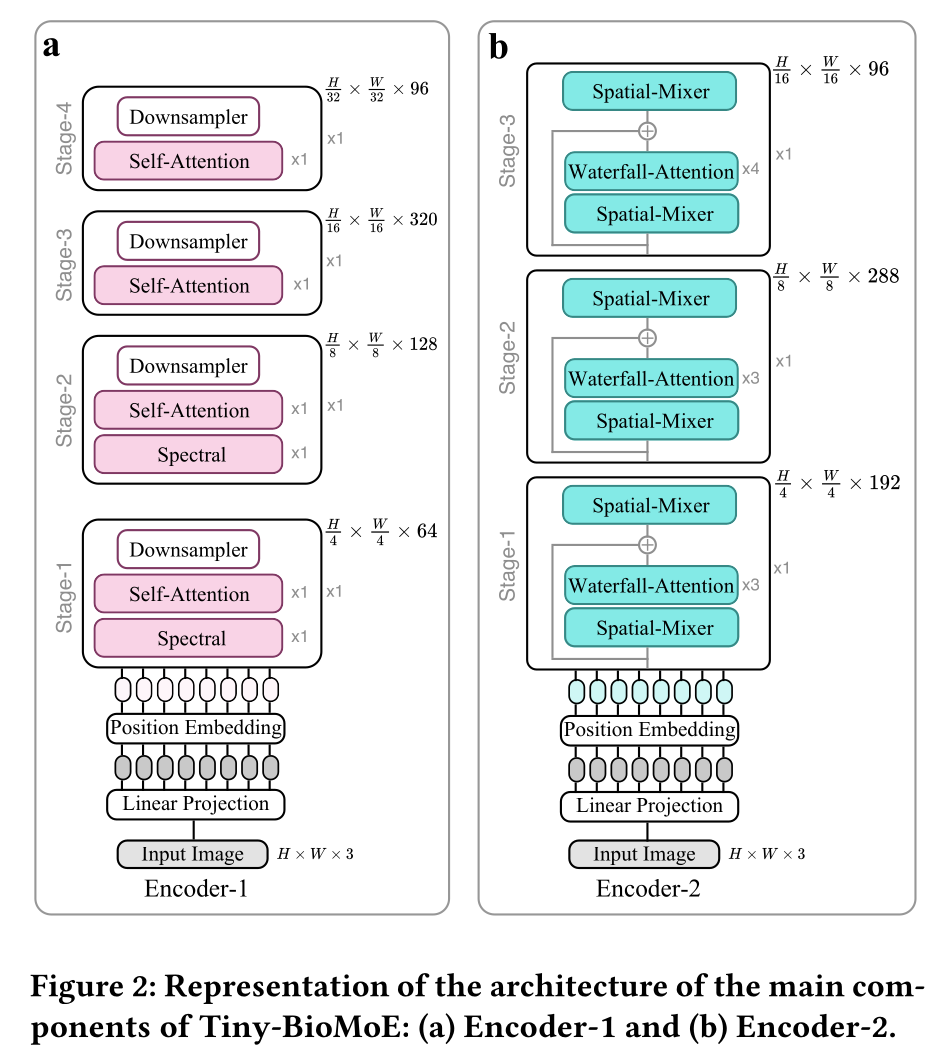
Tiny-BioMoE
a Lightweight Embedding Model for Biosignal Analysis
Tiny-BioMoE · 7.34 M parameters · 3.04 GFLOPs · 192-D embeddings · PyTorch ≥ 2.0
Paper
Tiny-BioMoE: a Lightweight Embedding Model for Biosignal Analysis
Highlights
| Feature | Description |
|---|---|
| Compact | <8 M parameters – runs comfortably on a laptop GPU / modern CPU |
| Cross-domain | Pre-trained on 4.4 M ECG, EMG & EEG representations via multi-task learning |
Table of Contents
Pre-trained Checkpoint
The checkpoint is stored under checkpoint/ in this repository.
| File | Size |
|---|---|
checkpoint/Tiny-BioMoE.pth |
89 MB |
Download options:
# direct file download
wget https://huggingface.co/stefanosgikas/TinyBioMoE/resolve/main/checkpoint/Tiny-BioMoE.pth
from huggingface_hub import hf_hub_download
ckpt_path = hf_hub_download(
repo_id="StefanosGkikas/Tiny-BioMoE",
filename="checkpoint/Tiny-BioMoE.pth"
)
print(ckpt_path)
Optional integrity check:
sha256sum checkpoint/Tiny-BioMoE.pth
The checkpoint contains:
model_state_dict # MoE backbone weights (SpectFormer-T-w + EfficientViT-w)
Quick start
Assumes PyTorch ≥ 2.0 and timm ≥ 0.9 are installed.
Repository layout expected:
.
├── docs/ # images for the model card
├── architecture/ # Python modules for the encoders / MoE
└── checkpoint/ # Tiny-BioMoE.pth
Extract embeddings
import torch, torch.nn as nn
from PIL import Image
from torchvision import transforms
from timm.models import create_model
# local "architecture" folder
from architecture import spectformer, efficientvit
emb_size, num_experts = 96, 2
final_emb_size = emb_size * num_experts # 192-D
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
class MoE(nn.Module):
def __init__(self, enc1, enc2):
super().__init__()
self.enc1, self.enc2 = enc1, enc2
self.ln_img = nn.LayerNorm((3, 224, 224))
self.ln_e = nn.LayerNorm(emb_size)
self.ln_out = nn.LayerNorm(final_emb_size)
self.fcn = nn.Sequential(nn.ELU(), nn.Linear(emb_size, emb_size),
nn.Hardtanh(0, 1))
@torch.no_grad()
def forward(self, x):
x = self.ln_img(x)
z1, *_ = self.enc1(x)
z2 = self.enc2(x)
z1 = self.ln_e(z1) * self.fcn(z1)
z2 = self.ln_e(z2) * self.fcn(z2)
return self.ln_out(torch.cat((z1, z2), 1))
enc1 = create_model('spectformer_t_w'); enc1.head = nn.Identity()
enc2 = create_model('EfficientViT_w'); enc2.head = nn.Identity()
backbone = MoE(enc1, enc2).to(device).eval()
state = torch.load("checkpoint/Tiny-BioMoE.pth", map_location=device)
backbone.load_state_dict(state['model_state_dict'])
tr = transforms.Compose([transforms.Resize((224,224)), transforms.ToTensor()])
img = Image.open('img.png').convert('RGB')
x = tr(img).unsqueeze(0).to(device)
feat = backbone(x).squeeze(0)
print(feat.shape)
Fine-tuning
import torch, torch.nn as nn
num_classes = 3
head = nn.Sequential(
nn.ELU(),
nn.Linear(192, num_classes)
)
for p in backbone.parameters():
p.requires_grad = False
model = nn.Sequential(backbone, head).to(device)
optimizer = torch.optim.AdamW(head.parameters(), lr=1e-3, weight_decay=1e-4)
Citation
@inproceedings{tiny_biomoe,
author = {Gkikas, Stefanos and Kyprakis, Ioannis and Tsiknakis, Manolis},
title = {Tiny-BioMoE: a Lightweight Embedding Model for Biosignal Analysis},
year = {2025},
isbn = {9798400720765},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3747327.3764788},
doi = {10.1145/3747327.3764788},
booktitle = {Companion Proceedings of the 27th International Conference on Multimodal Interaction},
pages = {117–126},
numpages = {10},
series = {ICMI '25 Companion}
}
Licence & acknowledgements
- Code & weights: MIT Licence – see
LICENSE.
Contact
Email Stefanos Gkikas: gkikas[at]ics[dot]forth[dot]gr / gikasstefanos[at]gmail[dot]com